Hydrogen suppresses cervical cancerScientific Research
Mechanism of hydrogen on cervical cancer suppression revealed by high‑throughput RNA sequencing
https://doi.org/10.3892/or.2021.8092
Xiaojun Liu, Zhijun Jin, Zhifeng Wang, Jianhong Dang, Guoqiang Chen, Lingling Li, Jing Wang, Jinghai Gao, Jing Chu
Abstract
Cervical cancer is considered one of the diseases with the highest mortality rate in women and limited treatment options. Hydrogen (H2) inhalation has been reported to have many tumor-inhibiting effects, but the exact mechanism remains unclear. In the present study, H2-treated HeLa cervical cancer cells and HaCaT keratinocytes and H2-inhaled HeLa xenograft mouse models were established. TUNEL, Cell Counting Kit-8, and Ki67 staining assays were used to detect apoptosis and proliferation. Oxidative stress was determined based on the levels of reactive oxygen species, malondialdehyde, and superoxide dismutase. Tumor growth was recorded every 3 days, and resected tumors were stained with hematoxylin and eosin. High-throughput RNA sequencing and subsequent Gene Ontology (GO) enrichment analysis was performed in HeLa-treated and untreated HeLa cells. The expression of hypoxia-inducible factor (HIF)-1α and NF-κB p65 was verified by western blot, immunohistochemistry and reverse transcription quantitative PCR. The results showed that in H2-treated HeLa cells, the rate of apoptosis was increased, and cell proliferation and oxidative stress were decreased, but not in HaCaT cells. Likewise, decreased tumor growth and cell proliferation and increased apoptosis were observed in H2-treated HeLa tumors.RNA-sequencing and GO analysis indicated that downregulation of HIF1A (HIF-1α mRNA) and RelA (NF-κB-p65) levels and reduced NF-κB signaling were associated with the antitumor activity of H2. Finally, decreased expression of HIF-1α and NF-κB p65 was observed at both transcriptional and translational levels in H2-treated HeLa cells and HeLa-derived tumors. In conclusion, this study demonstrates a novel mechanism of H2 against cervical cancer and may serve as a potential therapeutic target in clinical practice.
Results
H2 intervention reduces HeLa cell proliferation and promotes apoptosis and oxidative stress in a selective and dose-dependent manner
First, the effect of H2 treatment on HeLa cells was examined in vitro. As shown in Figure 1A and B, the proportion of TUNEL-positive cells was significantly increased in all H2-treated Hela cells compared with the control group (P<0.01). Treatment with 66.7% H2 significantly increased the rate of apoptosis. Importantly, 5 µg/mL cisplatin (IC50 = 12 µg/mL) treatment induced an increase in the rate of apoptosis in HeLa cells consistent with the effect of 66.7% H2 treatment. Notably, no significant difference in apoptosis rate was observed in HaCaT cells treated with or without hydrogen. The CCK-8 assay showed a gradual decrease in cell proliferation after treatment with 33.3% H2 and 66.7% H2, and further decreased proliferation of HeLa cells but not HaCaT cells over time (Fig. 1C). Furthermore, the effect of time was significantly associated with cell proliferation in HeLa cells and HaCaT cells. These results suggest that H2 has a specific concentration-dependent inhibitory effect on HeLa cells but not non-tumor HaCaT cells. Therefore, H2 was used for further testing at a concentration of 66.7%.
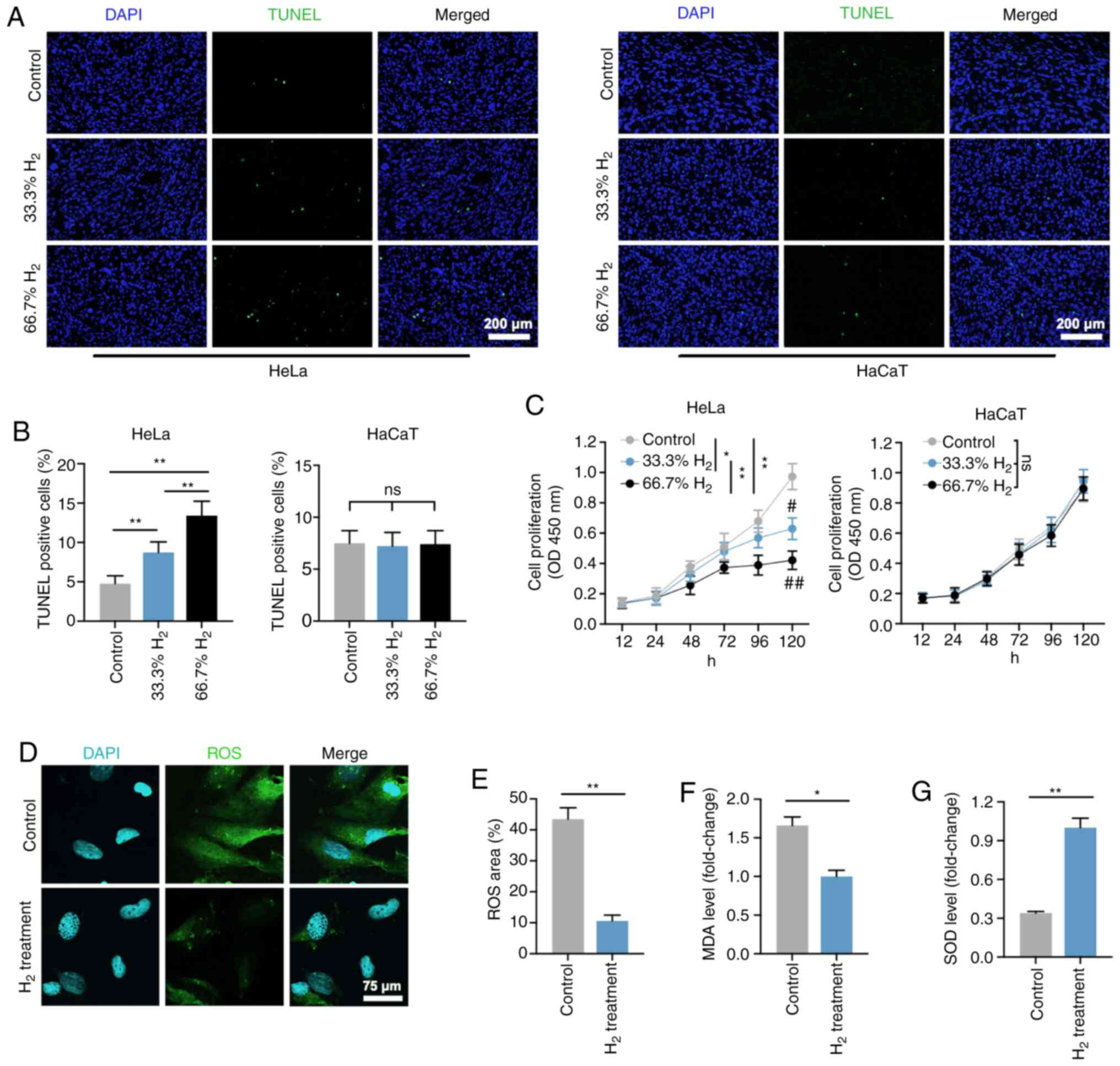
Figure 1. – Decreased cell proliferation, and increased cell death and oxidative stress level in H2-treated HeLa cervical cancer cells. (A) TUNEL staining of HeLa and HaCaT cells in the control, 33% H2 and 66.7% H2 groups. Furthermore, 5 µg/ml cisplatin treatment was used as a positive control (scale bar, 200 µm). (B) Statistical results of TUNEL-positive HeLa and HaCaT cells in the control, 33% H2 and 66.7% H2 groups (n=4 per group). **P<0.01, ns indicates no significant difference. (C) Cell proliferation curves of the control, 33% H2 and 66.7% H2 groups in HeLa and HaCaT cells during different time periods (n=3 per group). *P<0.05, **P<0.01 vs. indicated group, and #P<0.05, ##P<0.01 vs. control group at day 5, ns indicates no significant difference. (D) Fluorescent staining of ROS in control and 66.7% H2-treated HeLa cells (scale bar, 75 µm). (E) Statistical results of ROS area (%) in the control and 66.7% H2 groups (n=4 per group). **P<0.01. Statistical results of (F) malondialdehyde (MDA) and (G) superoxide dismutase (SOD) levels in the control and 66.7% H2 treatment groups (n=5 per group). *P<0.05, **P<0.01. H2, hydrogen; ROS, reactive oxygen species.
Aging mitochondria produce ROS due to overproduction of free radicals and reduced antioxidant defenses (26,27). The present study showed that ROS levels were significantly reduced in H2-treated HeLa cells compared with controls (P<0.01; Figure 1D and E), which is consistent with the previously reported antioxidative stress activity of H2 gas. 10). Furthermore, MDA levels, an indicator of lipid peroxidation, were also decreased in H2-treated HeLa cells (Fig. 1F), whereas in the H2 group, by contrast, elevated SOD levels, which are antioxidants, were observed An important representative of enzymes. Compared with the control group (P<0.01; Fig. 1G). These results suggest that H2 intervention induces HeLa cell apoptosis, inhibits cell proliferation, and reduces the level of oxidative stress.
Reduced tumor growth is observed in a H2-induced xenograft in vivo model
A total of 1 x 10 7 HeLa cells were injected subcutaneously in the left axilla of nude mice, and tumor volumes were recorded every 3 days starting from 7 days post-injection (Figure 2A). On days 31 and 34, the tumor volume of mice in the high H2 environment was significantly smaller than that of the control group (P<0.05 on day 31 and P<0.01 on day 34), consistent with the change in tumor weight on day 34 (P<0.01). 0.01; Figure 2B and C).
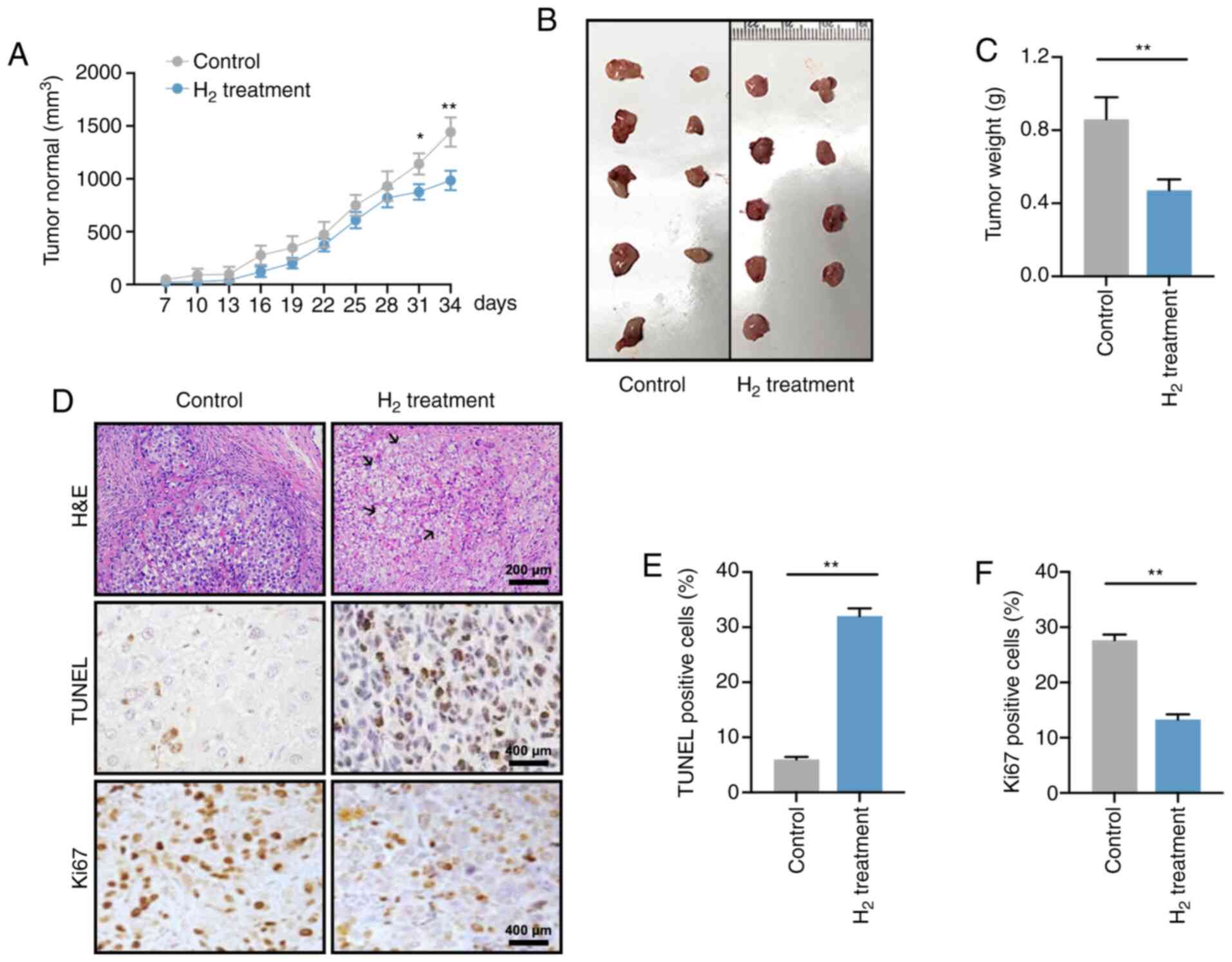
Figure 2. – H2 intervention reduces tumor growth in tumor-bearing mice. (A) Tumor volume curves from 7 to 34 days after cell injection in the control and H2 treatment tumor-bearing groups (n=9 per group). *P<0.05, **P<0.01. (B) Images of the excised tumors in control and H2 tumor-bearing mice. (C) Statistical results of tumor weight (units, g) in the control and H2 groups (n=9 per group). **P<0.01. (D) Hematoxylin and eosin staining (H&E) and immunohistochemistry for TUNEL and Ki67 staining in control tumor sections or sections after treatment with H2 (scale bar, 200 or 400 µm). Black arrows indicate nucleus fragmentation. Percentage of (E) TUNEL-positive cells or (F) Ki67-positive cells in control and H2-treated tumors (n=5 per group). **P<0.01. H2, hydrogen.
H&E staining showed increased tumor cell mortality in the H2 xenograft group with cell swelling and nuclear fragmentation and decay (Figure 2D; black arrows). Furthermore, H2 treatment significantly increased the apoptotic rate and decreased cell proliferation in HeLa cells (Figure 2E and F). Taken together, these data suggest that a high H2 environment suppresses tumor growth in tumor-bearing mice.
Bulk RNA sequencing of H2-treated HeLa cells and control cells
To investigate the underlying mechanism by which H2 inhibits tumor cell growth, whole-genome sequencing was performed (Figure 3). As shown in Figure S1A-C, 11 up-regulated and 16 down-regulated circular RNAs (circRNAs) were determined in H2-treated HeLa cells by the HiSeq4000 platform. In addition, 2 long non-coding RNAs (lncRNAs) were found to be up-regulated in H2-treated HeLa cells, while 6 lncRNAs were down-regulated (Fig. S2A-C). A total of 12 hsa microRNAs (miRNAs or miRs) were found to be up-regulated and 2 miRNAs were down-regulated, including hsa-miR-431-5p and hsa-miR-762 (Fig. S3A-C). Importantly, mRNA sequencing revealed that 10 mRNAs were up-regulated and 4 mRNAs were down-regulated in H2-treated HeLa cells, including HIF1A, miR23B, miR4651, and RelA (Figure 4A-C).miR23B and miR4651 are noncoding RNAs, while HIF1A and RelA are genes encoding the p65 subunit of HIF-1α and NF-κB, respectively. These results indicated that HIF-1α and NF-κB p65 subunit mRNA levels were reduced in H2-treated HeLa cells compared to controls.
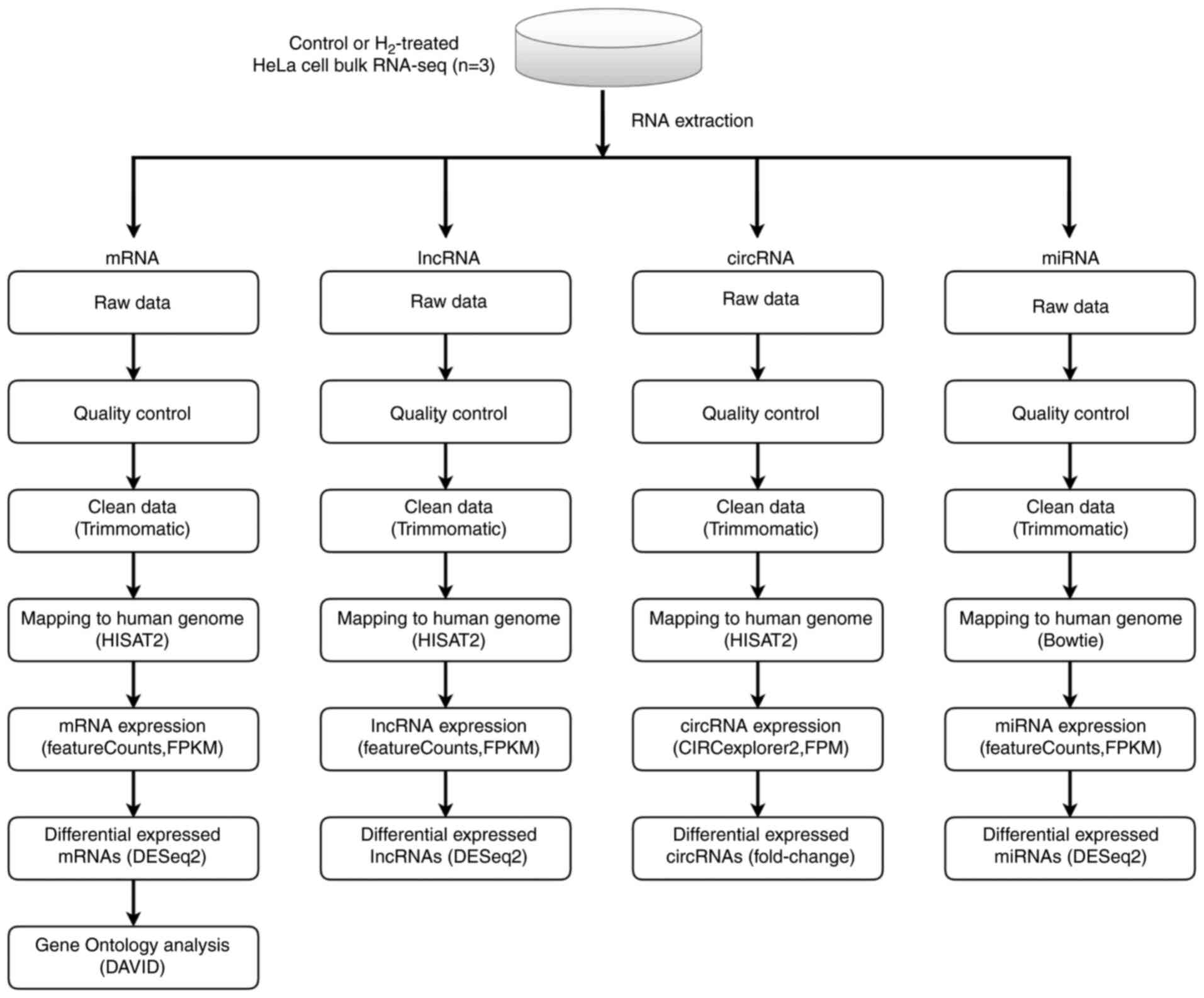
Figure 3. – Scheme of whole-genome (RNA) sequencing. RNA sequencing libraries were generated from 4 mg total RNA samples isolated from control or hydrogen-treated HeLa cells using TruSeq Stranded Total RNA Library Prep kit. Sequencing was carried out on Illumina NovaSeq 6000 and HiSeq X Ten platforms. After quality control, the clean reads of circular RNA, long non-coding RNA, microRNA and mRNA were then mapped to the human genome by HISAT2 software, and the analysis was performed using an DESeq2 software. Differential expression analysis was conducted with the DESeq2 platform (n=3 per group). Gene Ontology enrichment analysis of mRNA was performed using DAVID bioinformatics tool.
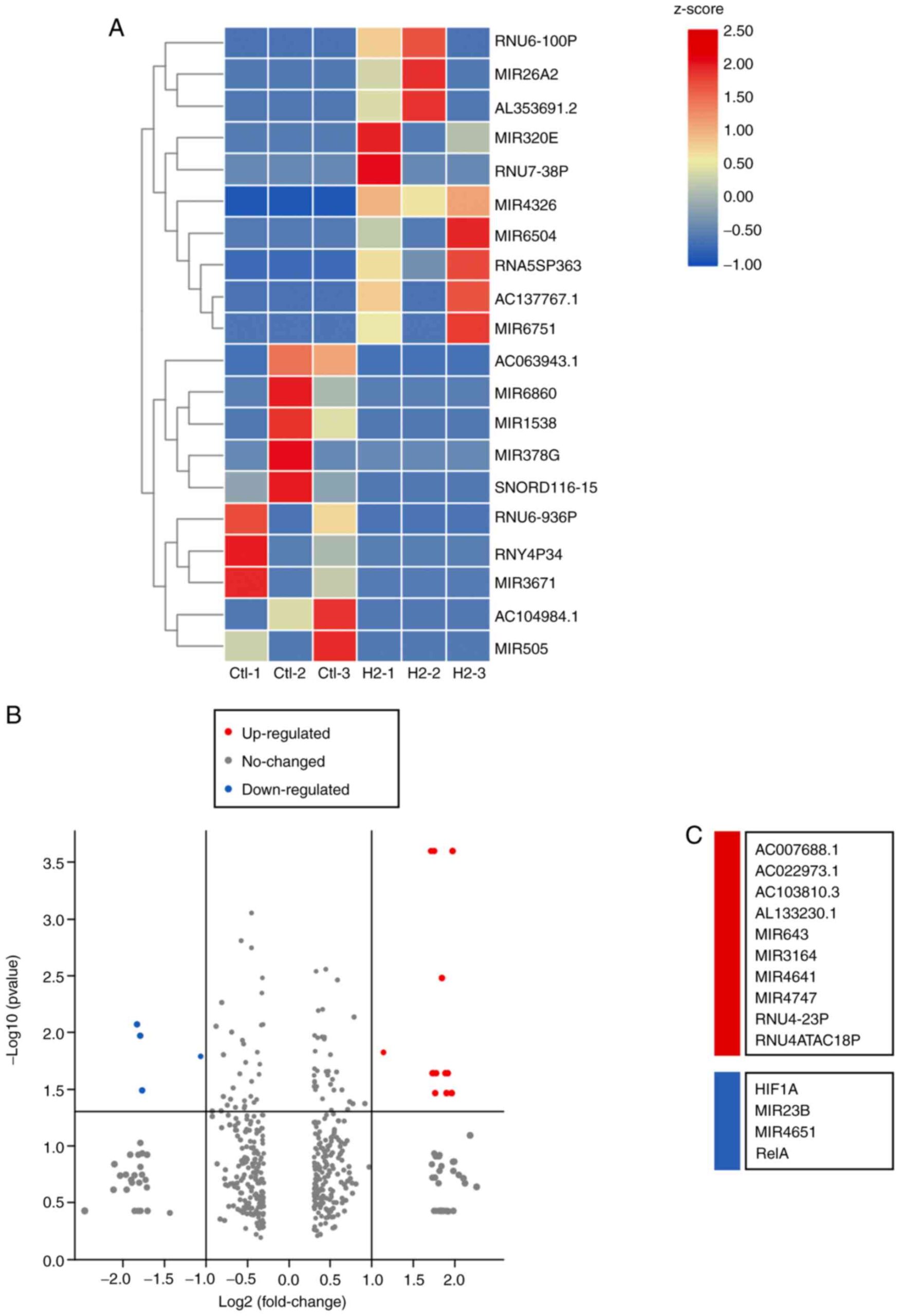
Figure 4. – Highly deregulated mRNAs in HeLa cells with or without H2 intervention. (A) A mRNA heatmap was generated based on the 10 maximum and 10 minimum FC values (n=3 per group). (B) Volcano plot for deregulated mRNAs [log2(FC) > 1 or < −1, adjusted P<0.05] between H2 and control groups. The area above the gray horizontal solid line indicates P<0.05. (C) Upregulated mRNAs (red) and downregulated mRNAs (aqua blue) are listed. H2, hydrogen; FC, fold-change.
GO enrichment analysis for HeLa cells with H2 intervention
To further confirm the above gene regulation in H2-treated HeLa cells, GO enrichment analysis was performed. As shown in Figure 5A, biological process (BP), cellular component (CC), and molecular function (MF) were analyzed. “Translation” and “NF-κB signaling” are the two most important processes in BP, and gene regulation mainly occurs in the nucleus and non-membrane-bound organelles. MF analysis showed that “DNA and RNA binding” predominated. The scatter plot of GO enrichment further indicated that the processes of “NF-κB signaling”, “protein transport” and “motility of multicellular organisms” were enriched in H2-treated HeLa cells (Fig. 5B). These results suggest that downregulation of NF-κB signaling may be an important target of H2 intervention in HeLa cells.
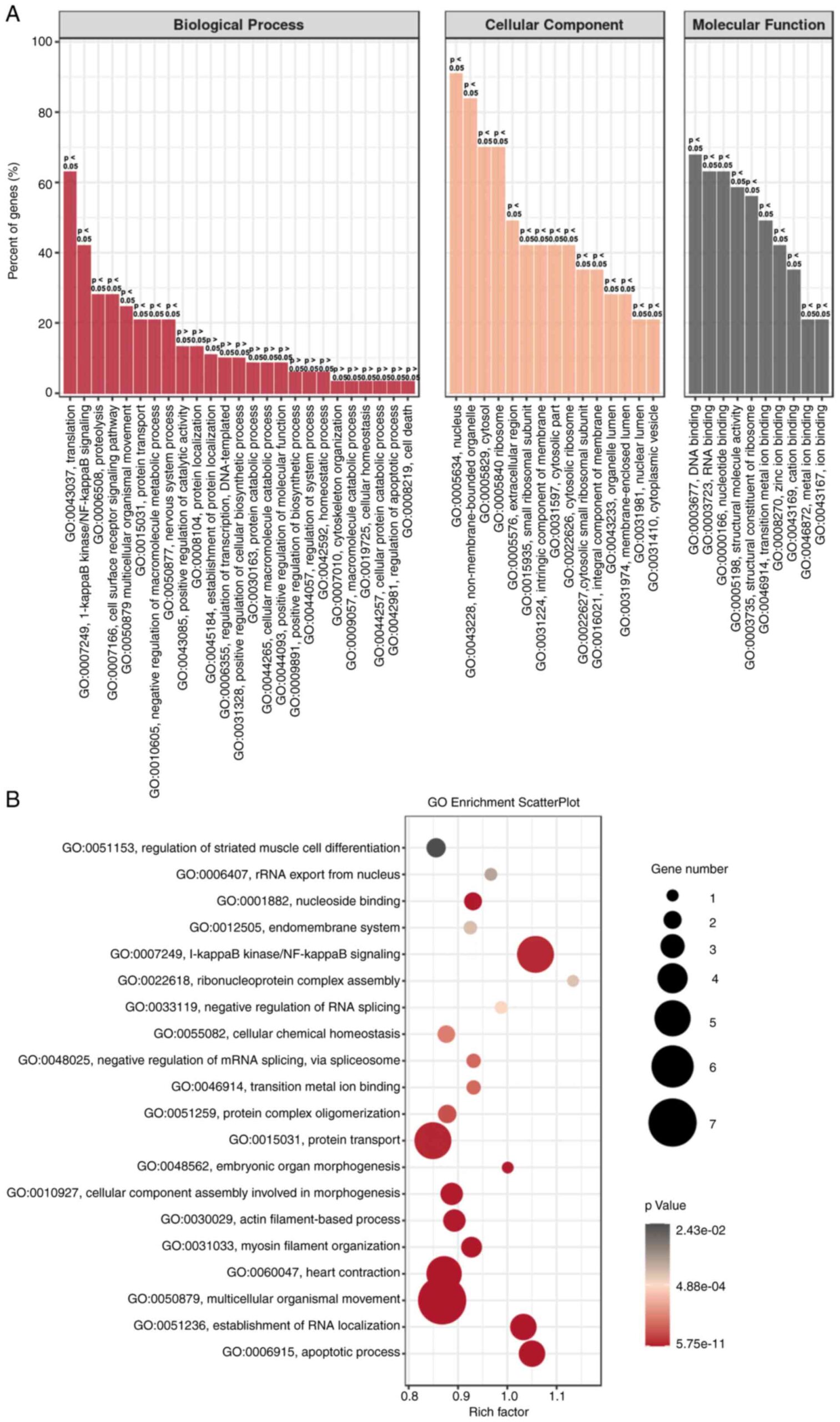
Figure 5. – GO enrichment analysis of H2-treated HeLa cells. (A) GO enrichment of BP, CC and MF analysis between H2-treated HeLa cells and controls. (B) GO enrichment scatterplot for BP, CC and MF progress between HeLa cells with or without H2 treatment. GO, Gene Ontology; BP, biological process, CC, cellular component; MF, molecular function; H2, hydrogen.
HIF-1α and NF-κB p65 expression levels are downregulated in H2-treated HeLa cells in vivo and in vitro.
Since the p65 subunit is the classically active type of the NF-κB family (28), this study validated the expression levels of NF-κB p65 and HIF-1α in HeLa cells. As expected, Western blot analysis indicated that HIF-1α and NF-κB p65 protein levels were significantly reduced in H2-treated HeLa cells compared with control HeLa cells (Fig. 6A-C). RT-qPCR analysis revealed that HIF-1α and NF-κB p65 (HIF1A and RelA) mRNA expression levels were increased in HeLa cells (Figure 6D and E).
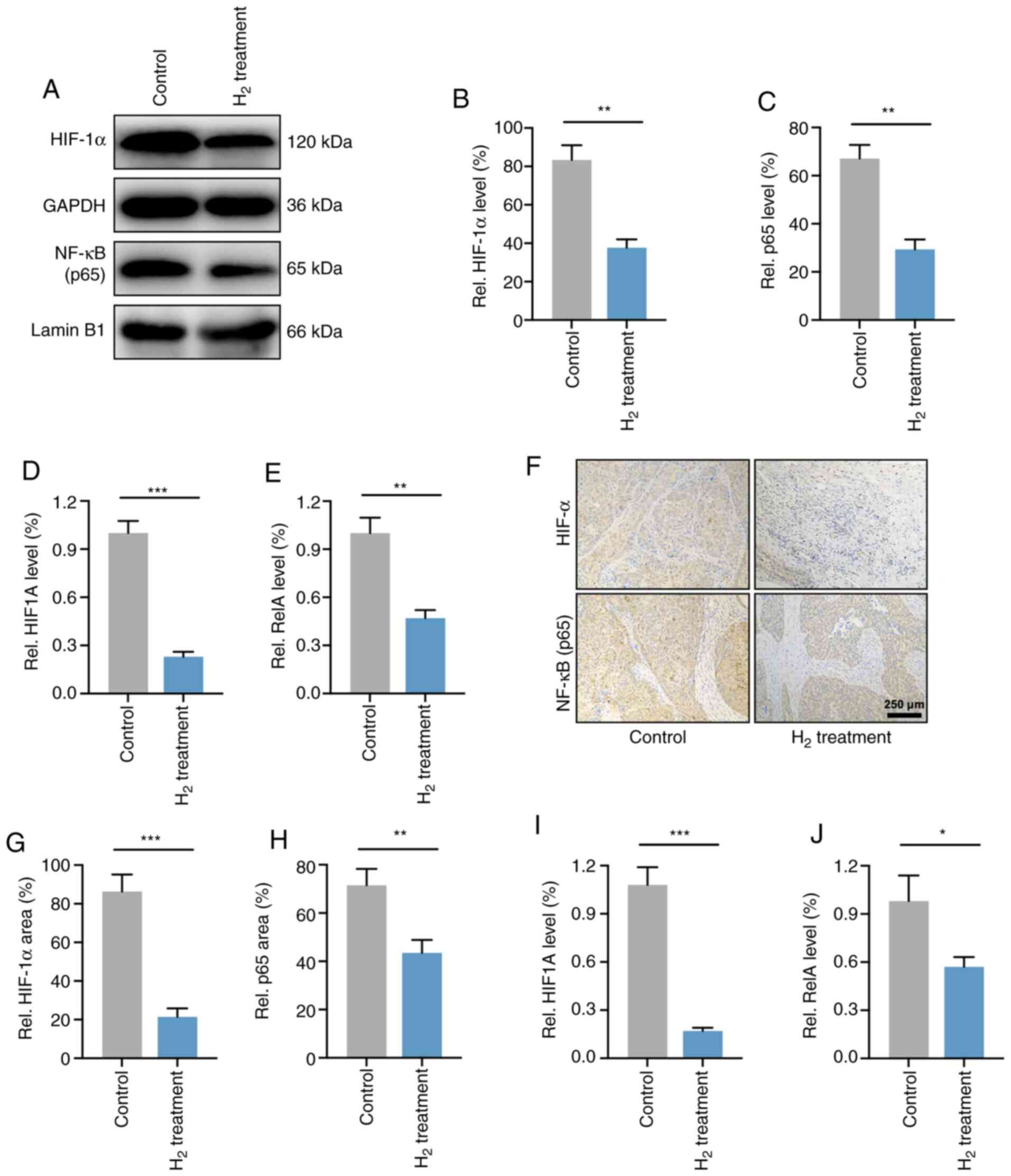
Figure 6. – H2 treatment downregulates both the gene and post-transcriptional levels of HIF-1α and NF-κB in HeLa cells and HeLa tumors. (A) Western blotting of HIF-1α and NF-κB in control and H2-treated HeLa cells. Cytoplasmic (GAPDH) and nuclear (lamin B1) loading controls were used. (B and C) Statistical results of (B) HIF-1α and (C) NF-κB protein expression levels in HeLa cells treated with H2 or not (n=4 per group). **P<0.01. RT-qPCR analysis for (D) HIF-1α mRNA (HIF1A) and (E) NF-κB mRNA (RelA) levels in control and H2-treated HeLa cells (n=4 per group). **P<0.01, ***P<0.001. (F) Immunohistochemical analysis for determining the HIF-1α and NF-κB expression in tumor-bearing mice after H2 intervention and their controls (scale bar, 250 µm). Percentage of positive area of (G) HIF-1α and (H) NF-κB in an in vivo xenograft model with or without H2 intervention (n=9 per group). **P<0.01, ***P<0.001. (I and J) RT-qPCR analysis for (I) HIF1A and (J) RelA expression in vivo (n=9 per group). *P<0.05, ***P<0.001. H2, hydrogen; HIF, hypoxia inducible factor; RT-qPCR, reverse transcription-quantitative PCR.
The levels of HIF-1α and NF-κB p65 were then determined in HeLa cell-derived tumors. As shown in Figure 6F-H, IHC showed that the expression of HIF-1α and NF-κB p65 was significantly reduced in H2-treated HeLa tumors compared with controls. In addition, mRNA levels were also examined in vivo and showed that HIF1A and RelA expression was significantly reduced in H2-treated tumors compared to control tumors (Figure 6I and J). These results indicated that the expression levels of HIF-1α and NF-κB p65 were downregulated in both HeLa cells and HeLa tumors.
References
Bray F, Ferlay J, Soerjomataram I, Siegel RL, Torre LA and Jemal A: Global cancer statistics 2018: GLOBOCAN estimates of incidence and mortality worldwide for 36 cancers in 185 countries. CA Cancer J Clin. 68:394–424. 2018. View Article : Google Scholar : PubMed/NCBI | |
Siegel RL, Miller KD and Jemal A: Cancer statistics, 2020. CA Cancer J Clin. 70:7–30. 2020. View Article : Google Scholar : PubMed/NCBI | |
Landoni F, Maneo A, Colombo A, Placa F, Milani R, Perego P, Favini G, Ferri L and Mangioni C: Randomised study of radical surgery versus radiotherapy for stage Ib-IIa cervical cancer. Lancet. 350:535–540. 1997. View Article : Google Scholar : PubMed/NCBI | |
Neeman E and Ben-Eliyahu S: Surgery and stress promote cancer metastasis: New outlooks on perioperative mediating mechanisms and immune involvement. Brain Behav Immun. 30 (Suppl 1):S32–S40. 2013. View Article : Google Scholar : PubMed/NCBI | |
Hodson R: Precision medicine. Nature. 537:S492016. View Article : Google Scholar : PubMed/NCBI | |
Tannock IF and Hickman JA: Limits to personalized cancer medicine. N Engl J Med. 375:1289–1294. 2016. View Article : Google Scholar : PubMed/NCBI | |
Sano M, Suzuki M, Homma K, Hayashida K, Tamura T, Matsuoka T, Katsumata Y, Onuki S and Sasaki J: Promising novel therapy with hydrogen gas for emergency and critical care medicine. Acute Med Surg. 5:113–118. 2017. View Article : Google Scholar : PubMed/NCBI | |
Ge L, Yang M, Yang NN, Yin XX and Song WG: Molecular hydrogen: A preventive and therapeutic medical gas for various diseases. Oncotarget. 8:102653–102673. 2017. View Article : Google Scholar : PubMed/NCBI | |
Ohta S: Molecular hydrogen as a preventive and therapeutic medical gas: Initiation, development and potential of hydrogen medicine. Pharmacol Ther. 144:1–11. 2014. View Article : Google Scholar : PubMed/NCBI | |
Tamura T, Suzuki M, Hayashida K, Kobayashi Y, Yoshizawa J, Shibusawa T, Sano M, Hori S and Sasaki J: Hydrogen gas inhalation alleviates oxidative stress in patients with post-cardiac arrest syndrome. J Clin Biochem Nutr. 67:214–221. 2020. View Article : Google Scholar : PubMed/NCBI | |
Chen L, Chao Y, Cheng P, Li N, Zheng H and Yang Y: UPLC-QTOF/MS-based metabolomics reveals the protective mechanism of hydrogen on mice with ischemic stroke. Neurochem Res. 44:1950–1963. 2019. View Article : Google Scholar : PubMed/NCBI | |
Yang Y, Liu PY, Bao W, Chen SJ, Wu FS and Zhu PY: Hydrogen inhibits endometrial cancer growth via a ROS/NLRP3/caspase-1/GSDMD-mediated pyroptotic pathway. BMC Cancer. 20:282020. View Article : Google Scholar : PubMed/NCBI | |
Wang D, Wang L, Zhang Y, Zhao Y and Chen G: Hydrogen gas inhibits lung cancer progression through targeting SMC3. Biomed Pharmacother. 104:788–797. 2018. View Article : Google Scholar : PubMed/NCBI | |
Harguindey S, Alfarouk K, Orozco JP, Hardonniere K, Stanciu D, Fais S and Devesa J: A new and integral approach to the etiopathogenesis and treatment of breast cancer based upon its hydrogen ion dynamics. Int J Mol Sci. 21:11102020. View Article : Google Scholar : PubMed/NCBI | |
Chen JB, Kong XF, Lv YY, Qin SC, Sun XJ, Mu F, Lu TY and Xu KC: ‘Real world survey’ of hydrogen-controlled cancer: A follow-up report of 82 advanced cancer patients. Med Gas Res. 9:115–121. 2019. View Article : Google Scholar : PubMed/NCBI | |
Andrews KR, Good JM, Miller MR, Luikart G and Hohenlohe PA: Harnessing the power of RADseq for ecological and evolutionary genomics. Nat Rev Genet. 17:81–92. 2016. View Article : Google Scholar : PubMed/NCBI | |
Masters JR: HeLa cells 50 years on: The good, the bad and the ugly. Nat Rev Cancer. 2:315–319. 2002. View Article : Google Scholar : PubMed/NCBI | |
Nagaraj N, Wisniewski JR, Geiger T, Cox J, Kircher M, Kelso J, Pääbo S and Mann M: Deep proteome and transcriptome mapping of a human cancer cell line. Mol Syst Biol. 7:5482011. View Article : Google Scholar : PubMed/NCBI | |
Tavakolian S, Goudarzi H, Eslami G and Faghihloo E: Transcriptional regulation of epithelial to mesenchymal transition related genes by lipopolysaccharide in human cervical cancer cell line HeLa. Asian Pac J Cancer Prev. 20:2455–2461. 2019. View Article : Google Scholar : PubMed/NCBI | |
Johrens K, Lazzerini L, Barinoff J, Sehouli J and Cichon G: Mesothelin as a target for cervical cancer therapy. Arch Gynecol Obstet. 299:211–216. 2019. View Article : Google Scholar : PubMed/NCBI | |
Lin Y, Ohkawara B, Ito M, Misawa N, Miyamoto K, Takegami Y, Masuda A, Toyokuni S and Ohno K: Molecular hydrogen suppresses activated Wnt/β-catenin signaling. Sci Rep. 6:319862016. View Article : Google Scholar : PubMed/NCBI | |
Kamalipooya S, Abdolmaleki P, Salemi Z, Javani Jouni F, Zafari J and Soleimani H: Simultaneous application of cisplatin and static magnetic field enhances oxidative stress in HeLa cell line. In Vitro Cell Dev Biol Anim. 53:783–790. 2017. View Article : Google Scholar : PubMed/NCBI | |
Metsalu T and Vilo J: ClustVis: A web tool for visualizing clustering of multivariate data using principal component analysis and heatmap. Nucleic Acids Res. 43:W566–W570. 2015. View Article : Google Scholar : PubMed/NCBI | |
Huang da W, Sherman BT and Lempicki RA: Systematic and integrative analysis of large gene lists using DAVID bioinformatics resources. Nat Protoc. 4:44–57. 2009. View Article : Google Scholar : PubMed/NCBI | |
Rao X, Huang X, Zhou Z and Lin X: An improvement of the 2^(-delta delta CT) method for quantitative real-time polymerase chain reaction data analysis. Biostat Bioinforma Biomath. 3:71–85. 2013.PubMed/NCBI | |
Finkel T and Holbrook NJ: Oxidants, oxidative stress and the biology of ageing. Nature. 408:239–247. 2000. View Article : Google Scholar : PubMed/NCBI | |
Costa V and Moradas-Ferreira P: Oxidative stress and signal transduction in Saccharomyces cerevisiae: Insights into ageing, apoptosis and diseases. Mol Aspects Med. 22:217–246. 2001. View Article : Google Scholar : PubMed/NCBI | |
DiDonato JA, Mercurio F and Karin M: NF-κB and the link between inflammation and cancer. Immunol Rev. 246:379–400. 2012. View Article : Google Scholar : PubMed/NCBI | |
Trottier H and Burchell AN: Epidemiology of mucosal human papillomavirus infection and associated diseases. Public Health Genomics. 12:291–307. 2009. View Article : Google Scholar : PubMed/NCBI | |
McBride AA: Oncogenic human papillomaviruses. Philos Trans R Soc Lond B Biol Sci. 372:201602732017. View Article : Google Scholar : PubMed/NCBI | |
Viarisio D, Gissmann L and Tommasino M: Human papillomaviruses and carcinogenesis: Well-established and novel models. Curr Opin Virol. 26:56–62. 2017. View Article : Google Scholar : PubMed/NCBI | |
St Laurent J, Luckett R and Feldman S: HPV vaccination and the effects on rates of HPV-related cancers. Curr Probl Cancer. 42:493–506. 2018. View Article : Google Scholar : PubMed/NCBI | |
Tornesello ML, Faraonio R, Buonaguro L, Annunziata C, Starita N, Cerasuolo A, Pezzuto F, Tornesello AL and Buonaguro FM: The role of microRNAs, long non-coding RNAs, and circular RNAs in cervical cancer. Front Oncol. 10:1502020. View Article : Google Scholar : PubMed/NCBI | |
Casarotto M, Fanetti G, Guerrieri R, Palazzari E, Lupato V, Steffan A, Polesel J, Boscolo-Rizzo P and Fratta E: Beyond microRNAs: Emerging role of other non-coding RNAs in HPV-driven cancers. Cancers (Basel). 12:12462020. View Article : Google Scholar : PubMed/NCBI | |
Wen X, Liu S, Sheng J and Cui M: Recent advances in the contribution of noncoding RNAs to cisplatin resistance in cervical cancer. PeerJ. 8:e92342020. View Article : Google Scholar : PubMed/NCBI | |
Vojtechova Z and Tachezy R: The role of miRNAs in virus-mediated oncogenesis. Int J Mol Sci. 19:12172018. View Article : Google Scholar : PubMed/NCBI | |
Calin GA and Croce CM: MicroRNA-cancer connection: The beginning of a new tale. Cancer Res. 66:7390–7394. 2006. View Article : Google Scholar : PubMed/NCBI | |
Gaur A, Jewell DA, Liang Y, Ridzon D, Moore JH, Chen C, Ambros VR and Israel MA: Characterization of microRNA expression levels and their biological correlates in human cancer cell lines. Cancer Res. 67:2456–2468. 2007. View Article : Google Scholar : PubMed/NCBI | |
Taniguchi K and Karin M: NF-κB, inflammation, immunity and cancer: Coming of age. Nat Rev Immunol. 18:309–324. 2018. View Article : Google Scholar : PubMed/NCBI | |
Hoesel B and Schmid JA: The complexity of NF-κB signaling in inflammation and cancer. Mol Cancer. 12:862013. View Article : Google Scholar : PubMed/NCBI | |
Guttridge DC, Albanese C, Reuther JY, Pestell RG and Baldwin AS Jr: NF-kappaB controls cell growth and differentiation through transcriptional regulation of cyclin D1. Mol Cell Biol. 19:5785–5799. 1999. View Article : Google Scholar : PubMed/NCBI | |
La Rosa FA, Pierce JW and Sonenshein GE: Differential regulation of the c-myc oncogene promoter by the NF-kappa B rel family of transcription factors. Mol Cell Biol. 14:1039–1044. 1994. View Article : Google Scholar : PubMed/NCBI | |
Perkins ND: Achieving transcriptional specificity with NF-kappa B. Int J Biochem Cell Biol. 29:1433–1448. 1997. View Article : Google Scholar : PubMed/NCBI | |
Vaupel P and Mayer A: Hypoxia in tumors: Pathogenesis-related classification, characterization of hypoxia subtypes, and associated biological and clinical implications. Adv Exp Med Biol. 812:19–24. 2014. View Article : Google Scholar : PubMed/NCBI | |
Zhou J, Schmid T, Schnitzer S and Brune B: Tumor hypoxia and cancer progression. Cancer Lett. 237:10–21. 2006. View Article : Google Scholar : PubMed/NCBI | |
Semenza GL: Oxygen sensing, hypoxia-inducible factors, and disease pathophysiology. Annu Rev Pathol. 9:47–71. 2014. View Article : Google Scholar : PubMed/NCBI | |
Wenger RH, Stiehl DP and Camenisch G: Integration of oxygen signaling at the consensus HRE. Sci STKE. 2005:re122015.PubMed/NCBI | |
Zhong H, De Marzo AM, Laughner E, Lim M, Hilton DA, Zagzag D, Buechler P, Isaacs WB, Semenza GL and Simons JW: Overexpression of hypoxia-inducible factor 1alpha in common human cancers and their metastases. Cancer Res. 59:5830–5835. 1999.PubMed/NCBI | |
Seeber LM, Horree N, Vooijs MA, Heintz AP, van der Wall E, Verheijen RH and van Diest PJ: The role of hypoxia inducible factor-1alpha in gynecological cancer. Crit Rev Oncol Hematol. 78:173–184. 2011. View Article : Google Scholar : PubMed/NCBI | |
Keith B, Johnson RS and Simon MC: HIF1α and HIF2α: Sibling rivalry in hypoxic tumour growth and progression. Nat Rev Cancer. 12:9–22. 2011. View Article : Google Scholar : PubMed/NCBI | |
Scholz CC and Taylor CT: Targeting the HIF pathway in inflammation and immunity. Curr Opin Pharmacol. 13:646–653. 2013. View Article : Google Scholar : PubMed/NCBI | |
Hervouet E, Cizkova A, Demont J, Vojtiskova A, Pecina P, Franssen-van Hal NL, Keijer J, Simonnet H, Ivanek R, Kmoch S, et al: HIF and reactive oxygen species regulate oxidative phosphorylation in cancer. Carcinogenesis. 29:1528–1537. 2008. View Article : Google Scholar : PubMed/NCBI | |
Hao Y, Yan Z, Zhang A, Hu S, Wang N, Luo XG, Ma W, Zhang TC and He H: IL-6/STAT3 mediates the HPV18 E6/E7 stimulated upregulation of MALAT1 gene in cervical cancer HeLa cells. Virus Res. 281:1979072020. View Article : Google Scholar : PubMed/NCBI |
DOI: 10.3892
Published on: 26/05/2023
Mechanism of hydrogen on cervical cancer suppression revealed by high‑throughput RNA sequencing
https://doi.org/10.3892/or.2021.8092
Xiaojun Liu, Zhijun Jin, Zhifeng Wang, Jianhong Dang, Guoqiang Chen, Lingling Li, Jing Wang, Jinghai Gao, Jing Chu
Abstract
Cervical cancer is considered one of the diseases with the highest mortality rate in women and limited treatment options. Hydrogen (H2) inhalation has been reported to have many tumor-inhibiting effects, but the exact mechanism remains unclear. In the present study, H2-treated HeLa cervical cancer cells and HaCaT keratinocytes and H2-inhaled HeLa xenograft mouse models were established. TUNEL, Cell Counting Kit-8, and Ki67 staining assays were used to detect apoptosis and proliferation. Oxidative stress was determined based on the levels of reactive oxygen species, malondialdehyde, and superoxide dismutase. Tumor growth was recorded every 3 days, and resected tumors were stained with hematoxylin and eosin. High-throughput RNA sequencing and subsequent Gene Ontology (GO) enrichment analysis was performed in HeLa-treated and untreated HeLa cells. The expression of hypoxia-inducible factor (HIF)-1α and NF-κB p65 was verified by western blot, immunohistochemistry and reverse transcription quantitative PCR. The results showed that in H2-treated HeLa cells, the rate of apoptosis was increased, and cell proliferation and oxidative stress were decreased, but not in HaCaT cells. Likewise, decreased tumor growth and cell proliferation and increased apoptosis were observed in H2-treated HeLa tumors.RNA-sequencing and GO analysis indicated that downregulation of HIF1A (HIF-1α mRNA) and RelA (NF-κB-p65) levels and reduced NF-κB signaling were associated with the antitumor activity of H2. Finally, decreased expression of HIF-1α and NF-κB p65 was observed at both transcriptional and translational levels in H2-treated HeLa cells and HeLa-derived tumors. In conclusion, this study demonstrates a novel mechanism of H2 against cervical cancer and may serve as a potential therapeutic target in clinical practice.
Results
H2 intervention reduces HeLa cell proliferation and promotes apoptosis and oxidative stress in a selective and dose-dependent manner
First, the effect of H2 treatment on HeLa cells was examined in vitro. As shown in Figure 1A and B, the proportion of TUNEL-positive cells was significantly increased in all H2-treated Hela cells compared with the control group (P<0.01). Treatment with 66.7% H2 significantly increased the rate of apoptosis. Importantly, 5 µg/mL cisplatin (IC50 = 12 µg/mL) treatment induced an increase in the rate of apoptosis in HeLa cells consistent with the effect of 66.7% H2 treatment. Notably, no significant difference in apoptosis rate was observed in HaCaT cells treated with or without hydrogen. The CCK-8 assay showed a gradual decrease in cell proliferation after treatment with 33.3% H2 and 66.7% H2, and further decreased proliferation of HeLa cells but not HaCaT cells over time (Fig. 1C). Furthermore, the effect of time was significantly associated with cell proliferation in HeLa cells and HaCaT cells. These results suggest that H2 has a specific concentration-dependent inhibitory effect on HeLa cells but not non-tumor HaCaT cells. Therefore, H2 was used for further testing at a concentration of 66.7%.
Figure 1. – Decreased cell proliferation, and increased cell death and oxidative stress level in H2-treated HeLa cervical cancer cells. (A) TUNEL staining of HeLa and HaCaT cells in the control, 33% H2 and 66.7% H2 groups. Furthermore, 5 µg/ml cisplatin treatment was used as a positive control (scale bar, 200 µm). (B) Statistical results of TUNEL-positive HeLa and HaCaT cells in the control, 33% H2 and 66.7% H2 groups (n=4 per group). **P<0.01, ns indicates no significant difference. (C) Cell proliferation curves of the control, 33% H2 and 66.7% H2 groups in HeLa and HaCaT cells during different time periods (n=3 per group). *P<0.05, **P<0.01 vs. indicated group, and #P<0.05, ##P<0.01 vs. control group at day 5, ns indicates no significant difference. (D) Fluorescent staining of ROS in control and 66.7% H2-treated HeLa cells (scale bar, 75 µm). (E) Statistical results of ROS area (%) in the control and 66.7% H2 groups (n=4 per group). **P<0.01. Statistical results of (F) malondialdehyde (MDA) and (G) superoxide dismutase (SOD) levels in the control and 66.7% H2 treatment groups (n=5 per group). *P<0.05, **P<0.01. H2, hydrogen; ROS, reactive oxygen species.
Aging mitochondria produce ROS due to overproduction of free radicals and reduced antioxidant defenses (26,27). The present study showed that ROS levels were significantly reduced in H2-treated HeLa cells compared with controls (P<0.01; Figure 1D and E), which is consistent with the previously reported antioxidative stress activity of H2 gas. 10). Furthermore, MDA levels, an indicator of lipid peroxidation, were also decreased in H2-treated HeLa cells (Fig. 1F), whereas in the H2 group, by contrast, elevated SOD levels, which are antioxidants, were observed An important representative of enzymes. Compared with the control group (P<0.01; Fig. 1G). These results suggest that H2 intervention induces HeLa cell apoptosis, inhibits cell proliferation, and reduces the level of oxidative stress.
Reduced tumor growth is observed in a H2-induced xenograft in vivo model
A total of 1 x 10 7 HeLa cells were injected subcutaneously in the left axilla of nude mice, and tumor volumes were recorded every 3 days starting from 7 days post-injection (Figure 2A). On days 31 and 34, the tumor volume of mice in the high H2 environment was significantly smaller than that of the control group (P<0.05 on day 31 and P<0.01 on day 34), consistent with the change in tumor weight on day 34 (P<0.01). 0.01; Figure 2B and C).
Figure 2. – H2 intervention reduces tumor growth in tumor-bearing mice. (A) Tumor volume curves from 7 to 34 days after cell injection in the control and H2 treatment tumor-bearing groups (n=9 per group). *P<0.05, **P<0.01. (B) Images of the excised tumors in control and H2 tumor-bearing mice. (C) Statistical results of tumor weight (units, g) in the control and H2 groups (n=9 per group). **P<0.01. (D) Hematoxylin and eosin staining (H&E) and immunohistochemistry for TUNEL and Ki67 staining in control tumor sections or sections after treatment with H2 (scale bar, 200 or 400 µm). Black arrows indicate nucleus fragmentation. Percentage of (E) TUNEL-positive cells or (F) Ki67-positive cells in control and H2-treated tumors (n=5 per group). **P<0.01. H2, hydrogen.
H&E staining showed increased tumor cell mortality in the H2 xenograft group with cell swelling and nuclear fragmentation and decay (Figure 2D; black arrows). Furthermore, H2 treatment significantly increased the apoptotic rate and decreased cell proliferation in HeLa cells (Figure 2E and F). Taken together, these data suggest that a high H2 environment suppresses tumor growth in tumor-bearing mice.
Bulk RNA sequencing of H2-treated HeLa cells and control cells
To investigate the underlying mechanism by which H2 inhibits tumor cell growth, whole-genome sequencing was performed (Figure 3). As shown in Figure S1A-C, 11 up-regulated and 16 down-regulated circular RNAs (circRNAs) were determined in H2-treated HeLa cells by the HiSeq4000 platform. In addition, 2 long non-coding RNAs (lncRNAs) were found to be up-regulated in H2-treated HeLa cells, while 6 lncRNAs were down-regulated (Fig. S2A-C). A total of 12 hsa microRNAs (miRNAs or miRs) were found to be up-regulated and 2 miRNAs were down-regulated, including hsa-miR-431-5p and hsa-miR-762 (Fig. S3A-C). Importantly, mRNA sequencing revealed that 10 mRNAs were up-regulated and 4 mRNAs were down-regulated in H2-treated HeLa cells, including HIF1A, miR23B, miR4651, and RelA (Figure 4A-C).miR23B and miR4651 are noncoding RNAs, while HIF1A and RelA are genes encoding the p65 subunit of HIF-1α and NF-κB, respectively. These results indicated that HIF-1α and NF-κB p65 subunit mRNA levels were reduced in H2-treated HeLa cells compared to controls.
Figure 3. – Scheme of whole-genome (RNA) sequencing. RNA sequencing libraries were generated from 4 mg total RNA samples isolated from control or hydrogen-treated HeLa cells using TruSeq Stranded Total RNA Library Prep kit. Sequencing was carried out on Illumina NovaSeq 6000 and HiSeq X Ten platforms. After quality control, the clean reads of circular RNA, long non-coding RNA, microRNA and mRNA were then mapped to the human genome by HISAT2 software, and the analysis was performed using an DESeq2 software. Differential expression analysis was conducted with the DESeq2 platform (n=3 per group). Gene Ontology enrichment analysis of mRNA was performed using DAVID bioinformatics tool.
Figure 4. – Highly deregulated mRNAs in HeLa cells with or without H2 intervention. (A) A mRNA heatmap was generated based on the 10 maximum and 10 minimum FC values (n=3 per group). (B) Volcano plot for deregulated mRNAs [log2(FC) > 1 or < −1, adjusted P<0.05] between H2 and control groups. The area above the gray horizontal solid line indicates P<0.05. (C) Upregulated mRNAs (red) and downregulated mRNAs (aqua blue) are listed. H2, hydrogen; FC, fold-change.
GO enrichment analysis for HeLa cells with H2 intervention
To further confirm the above gene regulation in H2-treated HeLa cells, GO enrichment analysis was performed. As shown in Figure 5A, biological process (BP), cellular component (CC), and molecular function (MF) were analyzed. “Translation” and “NF-κB signaling” are the two most important processes in BP, and gene regulation mainly occurs in the nucleus and non-membrane-bound organelles. MF analysis showed that “DNA and RNA binding” predominated. The scatter plot of GO enrichment further indicated that the processes of “NF-κB signaling”, “protein transport” and “motility of multicellular organisms” were enriched in H2-treated HeLa cells (Fig. 5B). These results suggest that downregulation of NF-κB signaling may be an important target of H2 intervention in HeLa cells.
Figure 5. – GO enrichment analysis of H2-treated HeLa cells. (A) GO enrichment of BP, CC and MF analysis between H2-treated HeLa cells and controls. (B) GO enrichment scatterplot for BP, CC and MF progress between HeLa cells with or without H2 treatment. GO, Gene Ontology; BP, biological process, CC, cellular component; MF, molecular function; H2, hydrogen.
HIF-1α and NF-κB p65 expression levels are downregulated in H2-treated HeLa cells in vivo and in vitro.
Since the p65 subunit is the classically active type of the NF-κB family (28), this study validated the expression levels of NF-κB p65 and HIF-1α in HeLa cells. As expected, Western blot analysis indicated that HIF-1α and NF-κB p65 protein levels were significantly reduced in H2-treated HeLa cells compared with control HeLa cells (Fig. 6A-C). RT-qPCR analysis revealed that HIF-1α and NF-κB p65 (HIF1A and RelA) mRNA expression levels were increased in HeLa cells (Figure 6D and E).
Figure 6. – H2 treatment downregulates both the gene and post-transcriptional levels of HIF-1α and NF-κB in HeLa cells and HeLa tumors. (A) Western blotting of HIF-1α and NF-κB in control and H2-treated HeLa cells. Cytoplasmic (GAPDH) and nuclear (lamin B1) loading controls were used. (B and C) Statistical results of (B) HIF-1α and (C) NF-κB protein expression levels in HeLa cells treated with H2 or not (n=4 per group). **P<0.01. RT-qPCR analysis for (D) HIF-1α mRNA (HIF1A) and (E) NF-κB mRNA (RelA) levels in control and H2-treated HeLa cells (n=4 per group). **P<0.01, ***P<0.001. (F) Immunohistochemical analysis for determining the HIF-1α and NF-κB expression in tumor-bearing mice after H2 intervention and their controls (scale bar, 250 µm). Percentage of positive area of (G) HIF-1α and (H) NF-κB in an in vivo xenograft model with or without H2 intervention (n=9 per group). **P<0.01, ***P<0.001. (I and J) RT-qPCR analysis for (I) HIF1A and (J) RelA expression in vivo (n=9 per group). *P<0.05, ***P<0.001. H2, hydrogen; HIF, hypoxia inducible factor; RT-qPCR, reverse transcription-quantitative PCR.
The levels of HIF-1α and NF-κB p65 were then determined in HeLa cell-derived tumors. As shown in Figure 6F-H, IHC showed that the expression of HIF-1α and NF-κB p65 was significantly reduced in H2-treated HeLa tumors compared with controls. In addition, mRNA levels were also examined in vivo and showed that HIF1A and RelA expression was significantly reduced in H2-treated tumors compared to control tumors (Figure 6I and J). These results indicated that the expression levels of HIF-1α and NF-κB p65 were downregulated in both HeLa cells and HeLa tumors.
References
Bray F, Ferlay J, Soerjomataram I, Siegel RL, Torre LA and Jemal A: Global cancer statistics 2018: GLOBOCAN estimates of incidence and mortality worldwide for 36 cancers in 185 countries. CA Cancer J Clin. 68:394–424. 2018. View Article : Google Scholar : PubMed/NCBI
Siegel RL, Miller KD and Jemal A: Cancer statistics, 2020. CA Cancer J Clin. 70:7–30. 2020. View Article : Google Scholar : PubMed/NCBI
Landoni F, Maneo A, Colombo A, Placa F, Milani R, Perego P, Favini G, Ferri L and Mangioni C: Randomised study of radical surgery versus radiotherapy for stage Ib-IIa cervical cancer. Lancet. 350:535–540. 1997. View Article : Google Scholar : PubMed/NCBI
Neeman E and Ben-Eliyahu S: Surgery and stress promote cancer metastasis: New outlooks on perioperative mediating mechanisms and immune involvement. Brain Behav Immun. 30 (Suppl 1):S32–S40. 2013. View Article : Google Scholar : PubMed/NCBI
Hodson R: Precision medicine. Nature. 537:S492016. View Article : Google Scholar : PubMed/NCBI
Tannock IF and Hickman JA: Limits to personalized cancer medicine. N Engl J Med. 375:1289–1294. 2016. View Article : Google Scholar : PubMed/NCBI
Sano M, Suzuki M, Homma K, Hayashida K, Tamura T, Matsuoka T, Katsumata Y, Onuki S and Sasaki J: Promising novel therapy with hydrogen gas for emergency and critical care medicine. Acute Med Surg. 5:113–118. 2017. View Article : Google Scholar : PubMed/NCBI
Ge L, Yang M, Yang NN, Yin XX and Song WG: Molecular hydrogen: A preventive and therapeutic medical gas for various diseases. Oncotarget. 8:102653–102673. 2017. View Article : Google Scholar : PubMed/NCBI
Ohta S: Molecular hydrogen as a preventive and therapeutic medical gas: Initiation, development and potential of hydrogen medicine. Pharmacol Ther. 144:1–11. 2014. View Article : Google Scholar : PubMed/NCBI
Tamura T, Suzuki M, Hayashida K, Kobayashi Y, Yoshizawa J, Shibusawa T, Sano M, Hori S and Sasaki J: Hydrogen gas inhalation alleviates oxidative stress in patients with post-cardiac arrest syndrome. J Clin Biochem Nutr. 67:214–221. 2020. View Article : Google Scholar : PubMed/NCBI
Chen L, Chao Y, Cheng P, Li N, Zheng H and Yang Y: UPLC-QTOF/MS-based metabolomics reveals the protective mechanism of hydrogen on mice with ischemic stroke. Neurochem Res. 44:1950–1963. 2019. View Article : Google Scholar : PubMed/NCBI
Yang Y, Liu PY, Bao W, Chen SJ, Wu FS and Zhu PY: Hydrogen inhibits endometrial cancer growth via a ROS/NLRP3/caspase-1/GSDMD-mediated pyroptotic pathway. BMC Cancer. 20:282020. View Article : Google Scholar : PubMed/NCBI
Wang D, Wang L, Zhang Y, Zhao Y and Chen G: Hydrogen gas inhibits lung cancer progression through targeting SMC3. Biomed Pharmacother. 104:788–797. 2018. View Article : Google Scholar : PubMed/NCBI
Harguindey S, Alfarouk K, Orozco JP, Hardonniere K, Stanciu D, Fais S and Devesa J: A new and integral approach to the etiopathogenesis and treatment of breast cancer based upon its hydrogen ion dynamics. Int J Mol Sci. 21:11102020. View Article : Google Scholar : PubMed/NCBI
Chen JB, Kong XF, Lv YY, Qin SC, Sun XJ, Mu F, Lu TY and Xu KC: ‘Real world survey’ of hydrogen-controlled cancer: A follow-up report of 82 advanced cancer patients. Med Gas Res. 9:115–121. 2019. View Article : Google Scholar : PubMed/NCBI
Andrews KR, Good JM, Miller MR, Luikart G and Hohenlohe PA: Harnessing the power of RADseq for ecological and evolutionary genomics. Nat Rev Genet. 17:81–92. 2016. View Article : Google Scholar : PubMed/NCBI
Masters JR: HeLa cells 50 years on: The good, the bad and the ugly. Nat Rev Cancer. 2:315–319. 2002. View Article : Google Scholar : PubMed/NCBI
Nagaraj N, Wisniewski JR, Geiger T, Cox J, Kircher M, Kelso J, Pääbo S and Mann M: Deep proteome and transcriptome mapping of a human cancer cell line. Mol Syst Biol. 7:5482011. View Article : Google Scholar : PubMed/NCBI
Tavakolian S, Goudarzi H, Eslami G and Faghihloo E: Transcriptional regulation of epithelial to mesenchymal transition related genes by lipopolysaccharide in human cervical cancer cell line HeLa. Asian Pac J Cancer Prev. 20:2455–2461. 2019. View Article : Google Scholar : PubMed/NCBI
Johrens K, Lazzerini L, Barinoff J, Sehouli J and Cichon G: Mesothelin as a target for cervical cancer therapy. Arch Gynecol Obstet. 299:211–216. 2019. View Article : Google Scholar : PubMed/NCBI
Lin Y, Ohkawara B, Ito M, Misawa N, Miyamoto K, Takegami Y, Masuda A, Toyokuni S and Ohno K: Molecular hydrogen suppresses activated Wnt/β-catenin signaling. Sci Rep. 6:319862016. View Article : Google Scholar : PubMed/NCBI
Kamalipooya S, Abdolmaleki P, Salemi Z, Javani Jouni F, Zafari J and Soleimani H: Simultaneous application of cisplatin and static magnetic field enhances oxidative stress in HeLa cell line. In Vitro Cell Dev Biol Anim. 53:783–790. 2017. View Article : Google Scholar : PubMed/NCBI
Metsalu T and Vilo J: ClustVis: A web tool for visualizing clustering of multivariate data using principal component analysis and heatmap. Nucleic Acids Res. 43:W566–W570. 2015. View Article : Google Scholar : PubMed/NCBI
Huang da W, Sherman BT and Lempicki RA: Systematic and integrative analysis of large gene lists using DAVID bioinformatics resources. Nat Protoc. 4:44–57. 2009. View Article : Google Scholar : PubMed/NCBI
Rao X, Huang X, Zhou Z and Lin X: An improvement of the 2^(-delta delta CT) method for quantitative real-time polymerase chain reaction data analysis. Biostat Bioinforma Biomath. 3:71–85. 2013.PubMed/NCBI
Finkel T and Holbrook NJ: Oxidants, oxidative stress and the biology of ageing. Nature. 408:239–247. 2000. View Article : Google Scholar : PubMed/NCBI
Costa V and Moradas-Ferreira P: Oxidative stress and signal transduction in Saccharomyces cerevisiae: Insights into ageing, apoptosis and diseases. Mol Aspects Med. 22:217–246. 2001. View Article : Google Scholar : PubMed/NCBI
DiDonato JA, Mercurio F and Karin M: NF-κB and the link between inflammation and cancer. Immunol Rev. 246:379–400. 2012. View Article : Google Scholar : PubMed/NCBI
Trottier H and Burchell AN: Epidemiology of mucosal human papillomavirus infection and associated diseases. Public Health Genomics. 12:291–307. 2009. View Article : Google Scholar : PubMed/NCBI
McBride AA: Oncogenic human papillomaviruses. Philos Trans R Soc Lond B Biol Sci. 372:201602732017. View Article : Google Scholar : PubMed/NCBI
Viarisio D, Gissmann L and Tommasino M: Human papillomaviruses and carcinogenesis: Well-established and novel models. Curr Opin Virol. 26:56–62. 2017. View Article : Google Scholar : PubMed/NCBI
St Laurent J, Luckett R and Feldman S: HPV vaccination and the effects on rates of HPV-related cancers. Curr Probl Cancer. 42:493–506. 2018. View Article : Google Scholar : PubMed/NCBI
Tornesello ML, Faraonio R, Buonaguro L, Annunziata C, Starita N, Cerasuolo A, Pezzuto F, Tornesello AL and Buonaguro FM: The role of microRNAs, long non-coding RNAs, and circular RNAs in cervical cancer. Front Oncol. 10:1502020. View Article : Google Scholar : PubMed/NCBI
Casarotto M, Fanetti G, Guerrieri R, Palazzari E, Lupato V, Steffan A, Polesel J, Boscolo-Rizzo P and Fratta E: Beyond microRNAs: Emerging role of other non-coding RNAs in HPV-driven cancers. Cancers (Basel). 12:12462020. View Article : Google Scholar : PubMed/NCBI
Wen X, Liu S, Sheng J and Cui M: Recent advances in the contribution of noncoding RNAs to cisplatin resistance in cervical cancer. PeerJ. 8:e92342020. View Article : Google Scholar : PubMed/NCBI
Vojtechova Z and Tachezy R: The role of miRNAs in virus-mediated oncogenesis. Int J Mol Sci. 19:12172018. View Article : Google Scholar : PubMed/NCBI
Calin GA and Croce CM: MicroRNA-cancer connection: The beginning of a new tale. Cancer Res. 66:7390–7394. 2006. View Article : Google Scholar : PubMed/NCBI
Gaur A, Jewell DA, Liang Y, Ridzon D, Moore JH, Chen C, Ambros VR and Israel MA: Characterization of microRNA expression levels and their biological correlates in human cancer cell lines. Cancer Res. 67:2456–2468. 2007. View Article : Google Scholar : PubMed/NCBI
Taniguchi K and Karin M: NF-κB, inflammation, immunity and cancer: Coming of age. Nat Rev Immunol. 18:309–324. 2018. View Article : Google Scholar : PubMed/NCBI
Hoesel B and Schmid JA: The complexity of NF-κB signaling in inflammation and cancer. Mol Cancer. 12:862013. View Article : Google Scholar : PubMed/NCBI
Guttridge DC, Albanese C, Reuther JY, Pestell RG and Baldwin AS Jr: NF-kappaB controls cell growth and differentiation through transcriptional regulation of cyclin D1. Mol Cell Biol. 19:5785–5799. 1999. View Article : Google Scholar : PubMed/NCBI
La Rosa FA, Pierce JW and Sonenshein GE: Differential regulation of the c-myc oncogene promoter by the NF-kappa B rel family of transcription factors. Mol Cell Biol. 14:1039–1044. 1994. View Article : Google Scholar : PubMed/NCBI
Perkins ND: Achieving transcriptional specificity with NF-kappa B. Int J Biochem Cell Biol. 29:1433–1448. 1997. View Article : Google Scholar : PubMed/NCBI
Vaupel P and Mayer A: Hypoxia in tumors: Pathogenesis-related classification, characterization of hypoxia subtypes, and associated biological and clinical implications. Adv Exp Med Biol. 812:19–24. 2014. View Article : Google Scholar : PubMed/NCBI
Zhou J, Schmid T, Schnitzer S and Brune B: Tumor hypoxia and cancer progression. Cancer Lett. 237:10–21. 2006. View Article : Google Scholar : PubMed/NCBI
Semenza GL: Oxygen sensing, hypoxia-inducible factors, and disease pathophysiology. Annu Rev Pathol. 9:47–71. 2014. View Article : Google Scholar : PubMed/NCBI
Wenger RH, Stiehl DP and Camenisch G: Integration of oxygen signaling at the consensus HRE. Sci STKE. 2005:re122015.PubMed/NCBI
Zhong H, De Marzo AM, Laughner E, Lim M, Hilton DA, Zagzag D, Buechler P, Isaacs WB, Semenza GL and Simons JW: Overexpression of hypoxia-inducible factor 1alpha in common human cancers and their metastases. Cancer Res. 59:5830–5835. 1999.PubMed/NCBI
Seeber LM, Horree N, Vooijs MA, Heintz AP, van der Wall E, Verheijen RH and van Diest PJ: The role of hypoxia inducible factor-1alpha in gynecological cancer. Crit Rev Oncol Hematol. 78:173–184. 2011. View Article : Google Scholar : PubMed/NCBI
Keith B, Johnson RS and Simon MC: HIF1α and HIF2α: Sibling rivalry in hypoxic tumour growth and progression. Nat Rev Cancer. 12:9–22. 2011. View Article : Google Scholar : PubMed/NCBI
Scholz CC and Taylor CT: Targeting the HIF pathway in inflammation and immunity. Curr Opin Pharmacol. 13:646–653. 2013. View Article : Google Scholar : PubMed/NCBI
Hervouet E, Cizkova A, Demont J, Vojtiskova A, Pecina P, Franssen-van Hal NL, Keijer J, Simonnet H, Ivanek R, Kmoch S, et al: HIF and reactive oxygen species regulate oxidative phosphorylation in cancer. Carcinogenesis. 29:1528–1537. 2008. View Article : Google Scholar : PubMed/NCBI
Hao Y, Yan Z, Zhang A, Hu S, Wang N, Luo XG, Ma W, Zhang TC and He H: IL-6/STAT3 mediates the HPV18 E6/E7 stimulated upregulation of MALAT1 gene in cervical cancer HeLa cells. Virus Res. 281:1979072020. View Article : Google Scholar : PubMed/NCBI



